NMX Research and Solutions
NMX is a partner contract research organization that is using their expertise in fragment-based ligand discovery and biophysics to launch and accelerate drug discovery programs. The following proof-of-concept project was designed to test if MFI’s tools could rank-order compounds and predict their binding poses in agreement with experimental data obtained by NMX.Furthermore, the study would give insight into how actionable information could be exchanged between MFI and NMX to discover hits and move them into leads more rationally and efficiently.
NMX derived 179 fragment analogs from the best hit of a broader fragment screen. They identified amino acids that showed changes in chemical shifts upon fragment binding to HRas. They further characterized the binding affinity of all fragment binders.
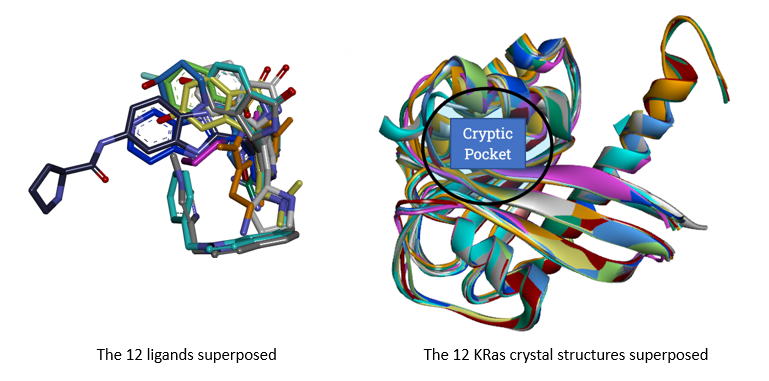
We performed standard sanity-check benchmarks of the structural models using FITTED (self- and cross-docking). The lack of available HRas crystal structures forced us to seek alternatives. The overall similarity between HRas and KRas is 89% over the entire sequence; the targeted cryptic pocket comprises identical amino acids. We opted to use KRas as a substitute for HRas. A set of 17 crystal structures were ultimately considered, among which 12 had ligands binding in the cryptic pocket (Figure 1). We selected one protein structure to use for virtual screening.
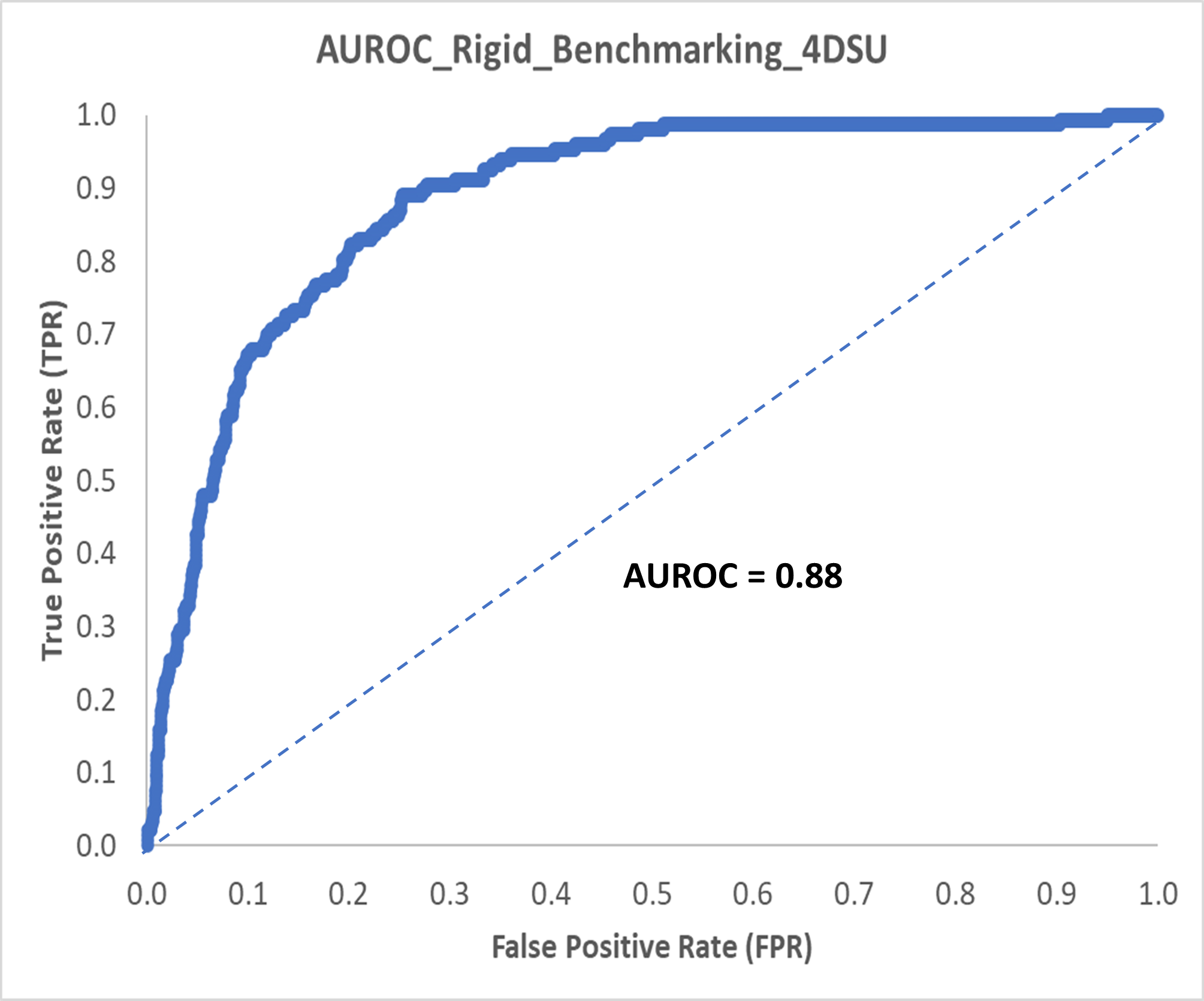
We bolstered the fragment set screened by NMX (approx. 150 actives) with generated decoys to create a total of 1500 compounds for our retrospective benchmarking. The AUROC of our retrospective benchmarking was 0.88 with good early enrichment factors: 20 out of the 37 actives were in the top 5% and all 6 actives in the top 1% were strong binders (Figure 2).
This collaborative project has highlighted the complementary nature of NMX’s and MFI’s expertise and the robustness of both team’s technology to kickstart projects. In such a study, the structural data provided by NMX plays a key role in supporting MFI’s predicted binding poses. In turn, by correctly posing and ranking compounds, MFI can rationally grow fragments into drug-like hits/leads.